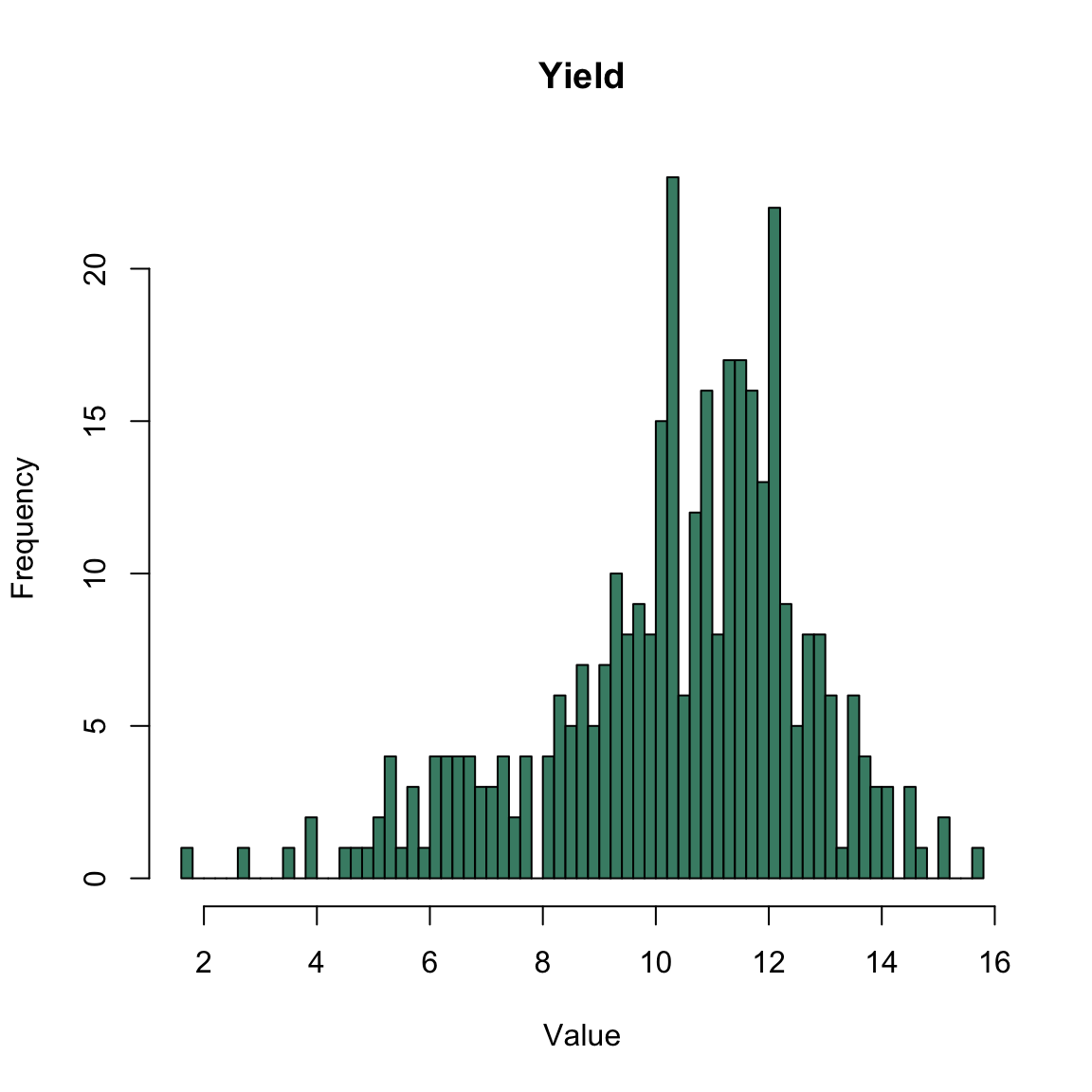
A maize experiment
A set of maize hybrids (N=345) crossed from male lines (M1, M2, M3, M4 and M5) and female lines (F1, F2, F3, F4, F5, F6).
The hybrids were planted in four environments, each with three replication.
Read in data
## Male Female ENV REP Yld Moist IVTD
## 1 M1 F1 2005_1 1 12.132546 11.80 70.01
## 2 M1 F1 2006_1 1 9.809719 11.16 70.48
## 3 M2 F1 2005_1 1 9.915348 11.70 69.71
## 4 M2 F1 2006_1 1 13.892623 11.10 67.07
## 5 M3 F1 2006_1 1 8.189426 11.60 70.46
## 6 M4 F1 2005_1 1 10.351882 11.50 69.67Across environments
To learn about how to do data visulization and other in R.
Many other R statistics resources
## Warning: package 'ggplot2' was built under R version 3.4.4corn$geno <- paste(corn$Male, corn$Female, sep="x")
fsize=18
p1 <- ggplot(data=corn, aes(x=REP, y=Yld, fill= as.factor(REP)) ) +
geom_violin() +
facet_wrap(~ ENV) +
#guides(fill=FALSE) + factor(trait, levels=out$trait)
labs(y=NULL, fill="Rep") +
xlab("") +
ylab("") +
theme_bw() +
theme(axis.text.x=element_blank(), axis.ticks.x=element_blank(),
axis.text=element_text(size=fsize),
axis.title=element_text(size=fsize, face="bold"),
legend.title = element_text(size=fsize, face="bold"),
legend.text = element_text(size=fsize))
p1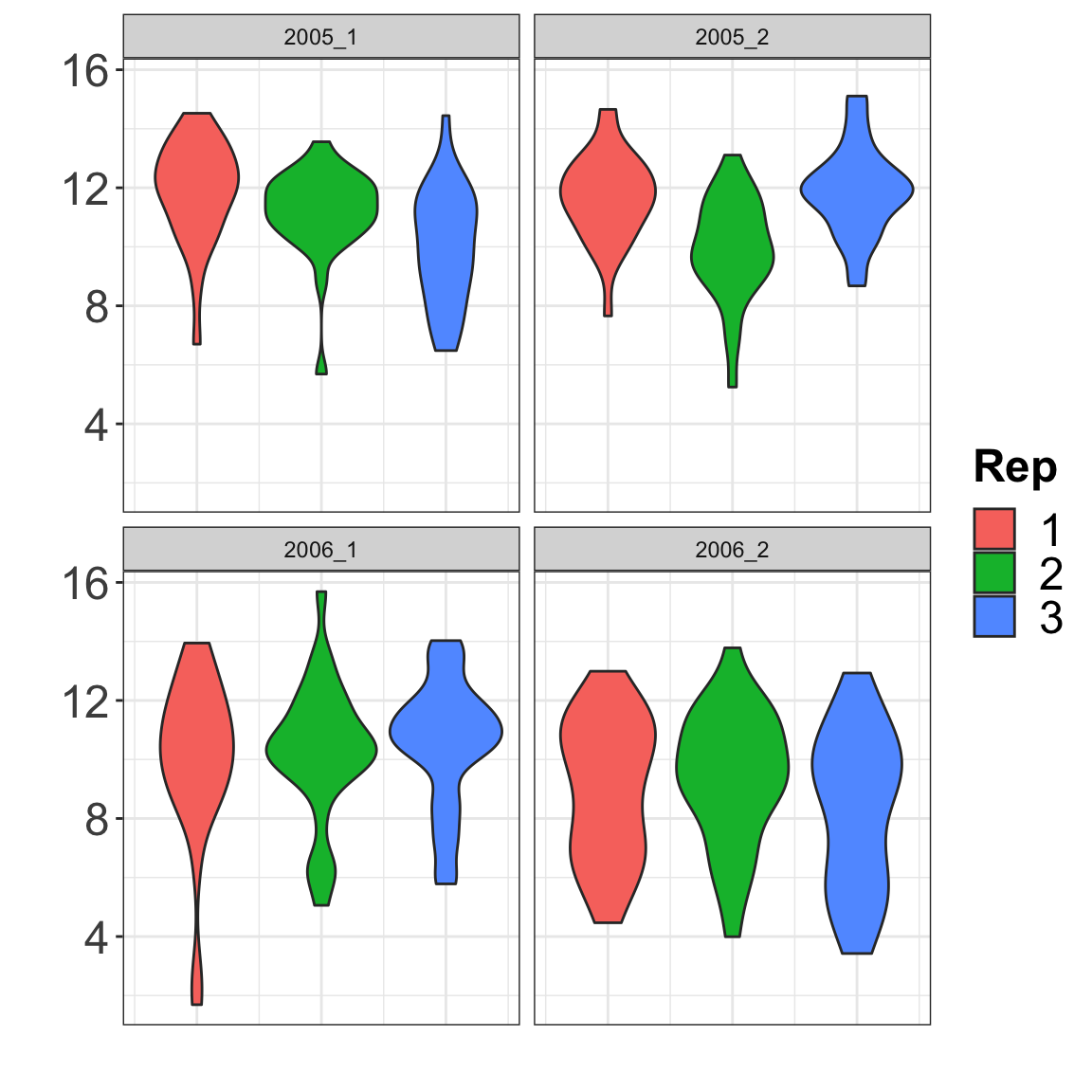
Half-sibs
One parent in common.
hs1 <- subset(corn, Male %in% c("M1", "M2"))
fit <- lm(Yld ~ Male + ENV + ENV:REP, data=hs1)
summary(aov(fit))## Df Sum Sq Mean Sq F value Pr(>F)
## Male 1 1.9 1.925 0.594 0.44232
## ENV 3 38.6 12.876 3.972 0.00949 **
## ENV:REP 4 25.0 6.238 1.925 0.10994
## Residuals 134 434.4 3.241
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1What is the among-families and within-families variance?
The intraclass correlation is 1.9/(1.9 + 3.2)
The covariance between two individuals
The covariance between two individuals in the same group is equal to the variance between groups.
Cov(HS) = Var(between)
Full-sibs
Both parents in common.
## Male Female ENV REP Yld Moist IVTD geno
## 1 M1 F1 2005_1 1 12.132546 11.80 70.01 M1xF1
## 2 M1 F1 2006_1 1 9.809719 11.16 70.48 M1xF1
## 12 M2 F2 2005_1 1 11.719874 11.80 72.24 M2xF2
## 13 M2 F2 2006_1 1 12.642069 11.70 67.30 M2xF2
## 58 M1 F1 2005_1 2 11.911893 11.60 71.33 M1xF1
## 59 M1 F1 2006_1 2 11.358482 11.40 67.92 M1xF1
## 71 M2 F2 2005_1 2 12.283671 11.90 71.18 M2xF2
## 72 M2 F2 2006_1 2 9.480686 11.90 65.50 M2xF2
## 116 M1 F1 2005_1 3 9.243844 11.80 71.04 M1xF1
## 117 M1 F1 2006_1 3 12.021751 11.40 70.10 M1xF1
## 128 M2 F2 2005_1 3 8.370777 12.30 68.33 M2xF2
## 129 M2 F2 2006_1 3 11.389737 11.80 67.83 M2xF2
## 172 M1 F1 2005_2 1 12.338039 12.10 71.37 M1xF1
## 173 M1 F1 2006_2 1 12.096800 9.60 64.92 M1xF1
## 184 M2 F2 2005_2 1 10.505628 11.60 67.45 M2xF2
## 185 M2 F2 2006_2 1 7.315236 9.10 65.67 M2xF2
## 230 M1 F1 2005_2 2 13.108079 11.90 65.39 M1xF1
## 231 M1 F1 2006_2 2 12.052070 9.30 64.85 M1xF1
## 242 M2 F2 2005_2 2 10.316791 11.80 66.76 M2xF2
## 243 M2 F2 2006_2 2 10.184848 9.30 61.35 M2xF2
## 288 M1 F1 2005_2 3 11.831043 12.20 68.05 M1xF1
## 289 M1 F1 2006_2 3 8.080784 9.10 65.01 M1xF1
## 300 M2 F2 2005_2 3 8.675930 11.70 65.54 M2xF2
## 301 M2 F2 2006_2 3 8.259983 9.00 64.05 M2xF2## Df Sum Sq Mean Sq F value Pr(>F)
## geno 1 9.18 9.176 4.061 0.0622 .
## ENV 3 8.93 2.976 1.317 0.3056
## ENV:REP 4 13.68 3.420 1.514 0.2482
## Residuals 15 33.89 2.259
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1Again, Cov(FS) = var(between)
Variance Components
corn$REP <- as.factor(corn$REP)
fit <- lm(Yld ~ Male + Female + ENV + ENV:REP, data=corn)
##Perform simple ANOVA
aovResult <- aov(fit)
##Extract table of mean squares
summary(aovResult)[[1]]## Df Sum Sq Mean Sq F value Pr(>F)
## Male 4 59.86 14.964 3.7421 0.005418 **
## Female 5 81.26 16.251 4.0641 0.001363 **
## ENV 3 285.33 95.112 23.7851 6.147e-14 ***
## ENV:REP 8 133.76 16.720 4.1813 8.843e-05 ***
## Residuals 324 1295.61 3.999
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1